Difference between revisions of "Ependymoma"
Jensflorian (talk | contribs) (→Microscopic: Supratentorial ependymoma) |
Jensflorian (talk | contribs) (→Molecular: Update) |
||
| Line 185: | Line 185: | ||
==Molecular== | ==Molecular== | ||
'''Supratentorial Ependymoma''' | |||
*SE, ZFTA-fusion positive: Adults and children (up to 80% of cases).<ref>{{Cite journal | last1 = Parker | first1 = M. | last2 = Mohankumar | first2 = KM. | last3 = Punchihewa | first3 = C. | last4 = Weinlich | first4 = R. | last5 = Dalton | first5 = JD. | last6 = Li | first6 = Y. | last7 = Lee | first7 = R. | last8 = Tatevossian | first8 = RG. | last9 = Phoenix | first9 = TN. | title = C11orf95-RELA fusions drive oncogenic NF-κB signalling in ependymoma. | journal = Nature | volume = 506 | issue = 7489 | pages = 451-5 | month = Feb | year = 2014 | doi = 10.1038/nature13109 | PMID = 24553141 }}</ref> | |||
**ZFTA-RELA fusion most common alteration. | |||
**Chromothripsis. | |||
**EPHB2 amplifications and CDKN2A deletions in a subset of these tumors<ref>{{Cite journal | last1 = Philip-Hollingsworth | first1 = S. | last2 = Hollingsworth | first2 = RI. | last3 = Dazzo | first3 = FB. | title = Host-range related structural features of the acidic extracellular polysaccharides of Rhizobium trifolii and Rhizobium leguminosarum. | journal = J Biol Chem | volume = 264 | issue = 3 | pages = 1461-6 | month = Jan | year = 1989 | doi = | PMID = 2912966 }}</ref> | |||
*SE, YAP-fusion positive. | |||
**Restricted to children (6-7% of all supratentorial ependymomas). | |||
**YAP-MAMLD fusion most common alteration. | |||
'''Posterior fossa Ependymoma''' | |||
Two distinct molecular subgroups exist in the posterior fossa:<ref>{{Cite journal | last1 = Witt | first1 = H. | last2 = Mack | first2 = SC. | last3 = Ryzhova | first3 = M. | last4 = Bender | first4 = S. | last5 = Sill | first5 = M. | last6 = Isserlin | first6 = R. | last7 = Benner | first7 = A. | last8 = Hielscher | first8 = T. | last9 = Milde | first9 = T. | title = Delineation of two clinically and molecularly distinct subgroups of posterior fossa ependymoma. | journal = Cancer Cell | volume = 20 | issue = 2 | pages = 143-57 | month = Aug | year = 2011 | doi = 10.1016/j.ccr.2011.07.007 | PMID = 21840481 }}</ref> | Two distinct molecular subgroups exist in the posterior fossa:<ref>{{Cite journal | last1 = Witt | first1 = H. | last2 = Mack | first2 = SC. | last3 = Ryzhova | first3 = M. | last4 = Bender | first4 = S. | last5 = Sill | first5 = M. | last6 = Isserlin | first6 = R. | last7 = Benner | first7 = A. | last8 = Hielscher | first8 = T. | last9 = Milde | first9 = T. | title = Delineation of two clinically and molecularly distinct subgroups of posterior fossa ependymoma. | journal = Cancer Cell | volume = 20 | issue = 2 | pages = 143-57 | month = Aug | year = 2011 | doi = 10.1016/j.ccr.2011.07.007 | PMID = 21840481 }}</ref> | ||
* Group A ependymomas: | * Group A ependymomas: | ||
| Line 198: | Line 207: | ||
**gene expression profiles similar to that of spinal cord ependymomas. | **gene expression profiles similar to that of spinal cord ependymomas. | ||
**increased Chromosomal 1q gains. <ref>{{Cite journal | last1 = Korshunov | first1 = A. | last2 = Witt | first2 = H. | last3 = Hielscher | first3 = T. | last4 = Benner | first4 = A. | last5 = Remke | first5 = M. | last6 = Ryzhova | first6 = M. | last7 = Milde | first7 = T. | last8 = Bender | first8 = S. | last9 = Wittmann | first9 = A. | title = Molecular staging of intracranial ependymoma in children and adults. | journal = J Clin Oncol | volume = 28 | issue = 19 | pages = 3182-90 | month = Jul | year = 2010 | doi = 10.1200/JCO.2009.27.3359 | PMID = 20516456 }}</ref> | **increased Chromosomal 1q gains. <ref>{{Cite journal | last1 = Korshunov | first1 = A. | last2 = Witt | first2 = H. | last3 = Hielscher | first3 = T. | last4 = Benner | first4 = A. | last5 = Remke | first5 = M. | last6 = Ryzhova | first6 = M. | last7 = Milde | first7 = T. | last8 = Bender | first8 = S. | last9 = Wittmann | first9 = A. | title = Molecular staging of intracranial ependymoma in children and adults. | journal = J Clin Oncol | volume = 28 | issue = 19 | pages = 3182-90 | month = Jul | year = 2010 | doi = 10.1200/JCO.2009.27.3359 | PMID = 20516456 }}</ref> | ||
==See also== | ==See also== | ||
Revision as of 12:50, 19 September 2022
| Ependymoma | |
|---|---|
| Diagnosis in short | |
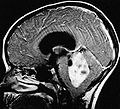
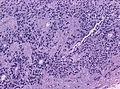
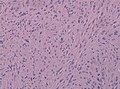
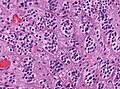
 Ependymoma grade II WHO. H&E stain | |
|
| |
| LM | Perivascular pseudorosettes, ependymal rosettes |
| Subtypes | Tanycytic, Clear cell, Papillary, Cellular |
| LM DDx | Subependymoma, Glioblastoma, Pilocytic astrocytoma, Oligodendroglioma |
| IHC | GFAP +ve |
| Prognosis | intermediate to poor (WHO Grades II & III) |
Ependymoma is a neuropathology tumour.
General
- Called the forgotten glial tumour.
- Anatomic location and molecular data is essential for tumor diagnosis.
Epidemiology:[1]
- Usual site:
- Adults: usually spinal cord.
- Children: usually posterior fossa.
- May be associated with neurofibromatosis type 2.
There are currently ten main ependymal tumors:[2]
- Supratentorial Subependymoma
- Supratentorial ependymoma, ZFTA-fusion positive
- Supratentorial ependymoma, YAP1-fusion positive
- Posterior fossa Subependymoma
- Posterior fossa ependymoma group A
- Posterior fossa ependymoma group B
- Spinal Subependymoma
- Spinal ependymoma
- Spinal ependymoma, MYCN-amplified
- Myxopapillary ependymoma
Ependymoma, NOS (not otherwise specified): Molecular analysis still missing. Ependymoma, NEC (not elsewhere classfied): Tumor cannot assigned to any of the defined entities.
Note: Molecularly defined ependymomas can be still graded as CNS grade 2 or 3 depending on histological features.
- Depreceated terminologies:
Gross
- Usually discrete and enhancing.
- Ventricular location, but also within the spinal cord.
- Dissemination possible.
- Myxopapillary ependymoma classically at filum terminale.
- Subependymoma typically seen in IVth ventricle.
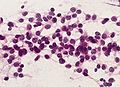
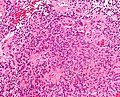
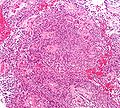
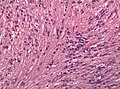
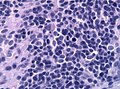
Microscopic
"Classic" ependymoma
- Come in two CNS WHO grades: 2 and 3.
- Usu. sharply demarcated from surrounding brain parenchyma.
Features:
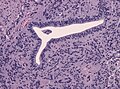
- Cells have a "tadpole-like" morphology.
- May also be described as ice cream cone-shaped.[5]
- Rosettes = circular nuclear free zones/cells arranged in a pseudoglandular fashion; comes in two flavours in ependymoma:
- Perivascular pseudorosettes = (tumour) cells arranged around a blood vessel; nuclei of cells distant from the blood vessel, i.e. rim of cytoplasm (from tumour cells) surround blood vessel (nucleus-free zone); more common than ependymal rosette... but less specific.
- Ependymal rosette (AKA true ependymal rosette) = rosette has an empty space at the centre - key feature.
- Nuclear features monotonous, i.e. "boring".[6]
- There is little variation in size, shape and staining.
- Hyalinized vessels.
- Calcification.
- Rare cases with cartilagineous metaplasia.[7]
- Branching capillaries usu. only in supratentorial ependymomas.
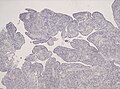
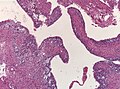
Supratentorial ependymoma
- Usu. connected to the ventricles.
- Mostly frontal or temporal lobe.
- Approx. 1/3 of all ependymal tumours (41% in children).
- Irregular CM enhancement.
- Cysts and/or calcification possible.
- Sharply demarcated from adjacent brain parenchyma.
- True ependymal rosettes are rare.
- Occasionally branching capillary vessels.
- Clear cell phenotypes more common than in other locations.
- Complete surgical resection is the best predictor.
- CSF spread in up to 15% of tumours.
DDx (supratentorial and posterior fossa ependymoma):
- Subependymoma.
- Glioblastoma (GBM).
- Gliomas with BCOR internal tandem duplication.
- Astroblastoma, MN1-altered.
- Invasive border = GBM; circumscribed border of lesion = ependymoma.
- Oligodendroglioma (Clear cell ependymoma))
- CNS embryonal tumour with BCOR internal tandem duplication.
DDx (spinal ependymoma):
- Pilocytic astrocytoma (Tanycytic ependymoma)
- Diffuse midline glioma, H3 K27-altered
- Small cell glioblastoma (MYCN-amplified spinal ependymoma)
Images
www:
- Ependymoma (flickr.com).
- Ependymoma - ependymal rosettes (ajnr.org).
- Anaplastic ependymoma - case 1 (upmc.edu).
- Anaplastic ependymoma - case 2 (upmc.edu).
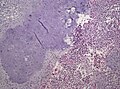
Tanycytic morphology in ependymoma must not confused with pilocytic astrocytoma. (WC/jensflorian)
Clear cell morphology in ependymoma may mimic oligodendroglioma. (WC/jensflorian)
Grading
Easy:
- Subependymoma = CNS WHO grade 1.
- Myxopapillary ependymoma = CNS WHO grade 2.
Not so easy: All other ependymomas: WHO CNS Grade 2 vs. Grade 3 depends on:
- Cellular density.
- Mitoses (no clear cut-off).
- Necrosis (not prognostic).
- Microvascular proliferation.
- Poor interobserver reliability[8]
Notes:
- Many tumours fall between grade 2 and grade 3.
- Rare cases with sarcomatous or cartilaginous components.[9][10]
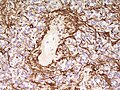
IHC
- Reticulin-ve.
- GFAP+ve.
- MIB1 (usu low).
- IDH-1-ve.
- EMA (dots and rings).[11]
- Widespread and strong EMA expression is indicative of YAP1-fused ependymoma.
- Olig2-ve.[12]
- H3K27me3 nuclear loss in Posterior fossa group A ependymoma (nuclear loss is diagnostic).[13]
- L1CAM in supratentorial tumors (expression indicates ZFTA fusion).[14]
- p65 nuclear +ve in ZFTA-fused ependymoma.
Molecular
Supratentorial Ependymoma
- SE, ZFTA-fusion positive: Adults and children (up to 80% of cases).[15]
- ZFTA-RELA fusion most common alteration.
- Chromothripsis.
- EPHB2 amplifications and CDKN2A deletions in a subset of these tumors[16]
- SE, YAP-fusion positive.
- Restricted to children (6-7% of all supratentorial ependymomas).
- YAP-MAMLD fusion most common alteration.
Posterior fossa Ependymoma Two distinct molecular subgroups exist in the posterior fossa:[17]
- Group A ependymomas:
- Group B ependymomas:
- typically adults.
- midline.
- relatively favorable clinical outcomes.
- gene expression profiles similar to that of spinal cord ependymomas.
- increased Chromosomal 1q gains. [20]
See also
References
- ↑ Kumar, Vinay; Abbas, Abul K.; Fausto, Nelson; Aster, Jon (2009). Robbins and Cotran pathologic basis of disease (8th ed.). Elsevier Saunders. pp. 1334. ISBN 978-1416031215.
- ↑ The International Agency for Research on Cancer (Editors: Louis, D.N.; Ohgaki, H.; Wiestler, O.D.; Cavenee, W.K.) (2007). Pathology and Genetics of Tumours of Tumors of the Central Nervous System (IARC WHO Classification of Tumours) (4th ed.). Lyon: World Health Organization. pp. 74. doi:10.1007/s00401-007-0243-4. ISBN 978-9283224303.
- ↑ Parker, M.; Mohankumar, KM.; Punchihewa, C.; Weinlich, R.; Dalton, JD.; Li, Y.; Lee, R.; Tatevossian, RG. et al. (Feb 2014). "C11orf95-RELA fusions drive oncogenic NF-κB signalling in ependymoma.". Nature 506 (7489): 451-5. doi:10.1038/nature13109. PMID 24553141.
- ↑ Pietsch, T.; Wohlers, I.; Goschzik, T.; Dreschmann, V.; Denkhaus, D.; Dörner, E.; Rahmann, S.; Klein-Hitpass, L. (Apr 2014). "Supratentorial ependymomas of childhood carry C11orf95-RELA fusions leading to pathological activation of the NF-κB signaling pathway.". Acta Neuropathol 127 (4): 609-11. doi:10.1007/s00401-014-1264-4. PMID 24562983.
- ↑ http://www.pathology.vcu.edu/WirSelfInst/tumor-2.html
- ↑ MUN. 6 Oct 2009.
- ↑ Wang, X.; Zhang, S.; Ye, Y.; Chen, Y.; Liu, X. (Jul 2012). "Ependymoma with cartilaginous metaplasia might have more aggressive behavior: a case report and literature review.". Brain Tumor Pathol 29 (3): 172-6. doi:10.1007/s10014-011-0079-4. PMID 22228122.
- ↑ Ellison, DW.; Kocak, M.; Figarella-Branger, D.; Felice, G.; Catherine, G.; Pietsch, T.; Frappaz, D.; Massimino, M. et al. (May 2011). "Histopathological grading of pediatric ependymoma: reproducibility and clinical relevance in European trial cohorts.". J Negat Results Biomed 10: 7. doi:10.1186/1477-5751-10-7. PMID 21627842.
- ↑ Vajtai, I.; Kuhlen, D.; Kappeler, A.; Mariani, L.; Zimmermann, A.; Paulus, W. (Jul 2010). "Rapid spontaneous malignant progression of supratentorial tanycytic ependymoma with sarcomatous features - "Ependymosarcoma".". Pathol Res Pract 206 (7): 493-8. doi:10.1016/j.prp.2009.07.013. PMID 19853384.
- ↑ Boukas, A.; Joshi, A.; Jenkins, A.; Holliman, D. (2013). "Extensive cartilaginous metaplasia of recurrent posterior fossa ependymoma: case report and review of the literature.". Pediatr Neurosurg 49 (2): 93-8. doi:10.1159/000356931. PMID 24401698.
- ↑ Hasselblatt, M.; Paulus, W. (Oct 2003). "Sensitivity and specificity of epithelial membrane antigen staining patterns in ependymomas.". Acta Neuropathol 106 (4): 385-8. doi:10.1007/s00401-003-0752-8. PMID 12898159.
- ↑ Švajdler, M.; Rychlý, B.; Mezencev, R.; Fröhlichová, L.; Bednárová, A.; Pataky, F.; Daum, O. (Jan 2016). "SOX10 and Olig2 as negative markers for the diagnosis of ependymomas: An immunohistochemical study of 98 glial tumors.". Histol Histopathol 31 (1): 95-102. doi:10.14670/HH-11-654. PMID 26287936.
- ↑ Panwalkar, P.; Clark, J.; Ramaswamy, V.; Hawes, D.; Yang, F.; Dunham, C.; Yip, S.; Hukin, J. et al. (Jul 2017). "Immunohistochemical analysis of H3K27me3 demonstrates global reduction in group-A childhood posterior fossa ependymoma and is a powerful predictor of outcome.". Acta Neuropathol. doi:10.1007/s00401-017-1752-4. PMID 28733933.
- ↑ Parker, M.; Mohankumar, KM.; Punchihewa, C.; Weinlich, R.; Dalton, JD.; Li, Y.; Lee, R.; Tatevossian, RG. et al. (Feb 2014). "C11orf95-RELA fusions drive oncogenic NF-κB signalling in ependymoma.". Nature 506 (7489): 451-5. doi:10.1038/nature13109. PMID 24553141.
- ↑ Parker, M.; Mohankumar, KM.; Punchihewa, C.; Weinlich, R.; Dalton, JD.; Li, Y.; Lee, R.; Tatevossian, RG. et al. (Feb 2014). "C11orf95-RELA fusions drive oncogenic NF-κB signalling in ependymoma.". Nature 506 (7489): 451-5. doi:10.1038/nature13109. PMID 24553141.
- ↑ Philip-Hollingsworth, S.; Hollingsworth, RI.; Dazzo, FB. (Jan 1989). "Host-range related structural features of the acidic extracellular polysaccharides of Rhizobium trifolii and Rhizobium leguminosarum.". J Biol Chem 264 (3): 1461-6. PMID 2912966.
- ↑ Witt, H.; Mack, SC.; Ryzhova, M.; Bender, S.; Sill, M.; Isserlin, R.; Benner, A.; Hielscher, T. et al. (Aug 2011). "Delineation of two clinically and molecularly distinct subgroups of posterior fossa ependymoma.". Cancer Cell 20 (2): 143-57. doi:10.1016/j.ccr.2011.07.007. PMID 21840481.
- ↑ Mack, SC.; Witt, H.; Piro, RM.; Gu, L.; Zuyderduyn, S.; Stütz, AM.; Wang, X.; Gallo, M. et al. (Feb 2014). "Epigenomic alterations define lethal CIMP-positive ependymomas of infancy.". Nature 506 (7489): 445-50. doi:10.1038/nature13108. PMID 24553142.
- ↑ Panwalkar, P.; Clark, J.; Ramaswamy, V.; Hawes, D.; Yang, F.; Dunham, C.; Yip, S.; Hukin, J. et al. (Jul 2017). "Immunohistochemical analysis of H3K27me3 demonstrates global reduction in group-A childhood posterior fossa ependymoma and is a powerful predictor of outcome.". Acta Neuropathol. doi:10.1007/s00401-017-1752-4. PMID 28733933.
- ↑ Korshunov, A.; Witt, H.; Hielscher, T.; Benner, A.; Remke, M.; Ryzhova, M.; Milde, T.; Bender, S. et al. (Jul 2010). "Molecular staging of intracranial ependymoma in children and adults.". J Clin Oncol 28 (19): 3182-90. doi:10.1200/JCO.2009.27.3359. PMID 20516456.